NGS Adapter, an essential part of the next-generation sequencing library, plays a role in connecting the tested DNA fragment and the Flow cell (Sequencing chip). The efficiency of the joint is an important factor in determining the quality and yield of the library. So what is an NGS adapter? What are the common types of NGS adapters? And how to choose the right NGS adapters for your sequencing platforms?
2. What factors should you consider when choosing the NGS index?
3. What are the common types of indexes?
4. What are the common types of NGS adapters?
5. How to choose the right NGS adapters for your sequencing platforms?
1. What is NGS Adapter?
NGS Adapter, a series of adapters in sequencing, is a short nucleotide sequence with a known sequence. It is ligated to both ends of the target nucleic acid fragment. During sequencing, it starts sequencing by hybridizing with the known sequence on the Flow cell to combine the library to the chip. So what is the structure of the NGS adapter?
Taking the Illumina platform as an example, an NGS adapter can be divided into three parts:
P5 and P7: Sequence combined with P5 and P7 ends on the Flow cell to fix the library on the sequencing chip, facilitating cluster reaction through Bridge-PCR.
Rd1 SP and Rd2 SP (Read1/Read2 sequencing primer): Binding regions of sequencing primers, indicating the position where the sequence begins to be read.
Index (also known as barcode): a known synthetic sequence used to distinguish different samples in the sequencing of the mixed library.
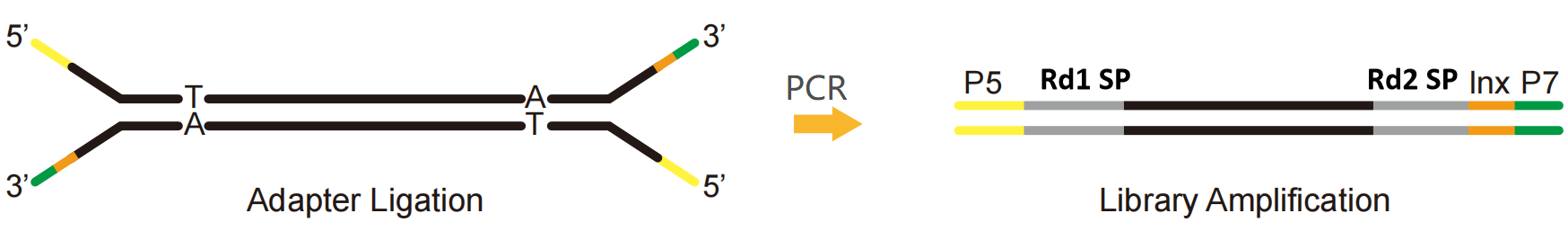
Fig. 1 Illumina platform single index library

Fig. 2 MGI platform single-end index library
With the increase in sequencing throughput, multiple samples can be sequenced at the same time. So how to distinguish diverse samples is especially important. As mentioned before, index sequences of NGS adapter are used to differentiate various samples in next-generation sequencing (NGS). What factors should you consider when choosing the index? Please continue to read...
2. What factors should you consider when choosing index?
The index is generally at the length of 6nt-18nt and is divided into a single index and a double index according to the number of indexes. The double index is located at both ends of the fragment to be tested. Base balance and fluorescence balance should be considered when selecting an index combination.
Base balance refers to the balance between multiple indexes, rather than the base balance in a single index. It needs to be considered from both base types and base distribution. The principle of combination is that the four bases A/T/C/G in the same group of indexes need to be included, and the proportion of these four bases is close, accounting for about 25% respectively.
Fluorescence signal balance refers to the choice to ensure the balance of fluorescent signals when the base balance cannot be guaranteed. In the 4-channel sequencer in the Illumina platform, dG/dT is labeled with green fluorescence, and dC/dA is labeled with red fluorescence. During sequencing, both green and red fluorescence signals must exist in each cycle to ensure successful sequencing. Therefore, the balance between the green signal and the red signal should be considered when selecting the index.
3. What are the common types of indexes?
The common dual indexes usually include Unique Dual Index (UDI), Unique Dual Barcode (UDB), and Combined Dual Index (CDI), which significantly reduce index hopping and misassignment.
UDI&UDB: the indexes at both ends are one-to-one corresponding, designed in groups, and can be cross-verified at both ends;
A Stubby UDI Primer Kit for Illumina supplied by Yeasen (Cat#12404ES/12405ES)>>
CDI: the indexes at both ends can be combined according to certain requirements to form a double-ended index library;
A 384 CDI Primer for Illumina,Set1-Set2 supplied by Yeasen (Cat#12412ES/12413ES)>>
In order to improve the throughput and amplification efficiency and reduce the sequencing cost, Illumina introduced the array flow cell (PFCT) and exclusive amplification (ExAmp) clustering technology for Novaseq and other high-throughput sequencers but inadvertently amplified the sample label mismatch phenomenon and index hopping.

Fig. 3 Illumina different instrument models adopt non-patterned flow cell or patterned flow cell
In order to make up for the index hooping problem highlighted by the sequencing platforms such as HiSeq3000/4000, HiSeq X Series, and NovaSeq, Illumina proposed the strategy of putting the index on both ends of the library, which can perform bilateral verification and eliminate the mismatched adapters. When using the unique indexes at both ends, the index error allocation rate will be reduced to 0.01%. Compared with the previous conventional index permutation group combination method, index hopping will be reduced by two orders of magnitude.
In the construction of a PCR-free library, a single-end index adapter is available. The label mismatch is mainly caused by sequencing errors. Overall, the label mismatch rate is low (average 0.0004%, up to 0.001%). However, in the construction of a targeted capture library, the crosstalk problem is amplified because multiple steps will lead to label mismatch and the UDI/UDB/CDI adapters are usually used.
4.What are the common types of NGS adapters?
With the development of sequencing technology, there are more and more types of adapters, such as single index/double index adapters (as mentioned in section 3), UMI adapters, transposase adapters, complete/incomplete adapters, etc., which are suitable for a variety of application scenarios. This part systematically sorts out these adapters to give you the foundation of adapter selection.
4.1 UMI Adapter
Unique molecular identifier(UMI) adapter is an edge tool for low-frequency mutation detection and absolute quantification. UMI is a random synthetic sequence with a known sequence. It can be designed as a completely random nucleotide chain, partial degenerate nucleotide chain, or fixed nucleotide chain. The length is usually 10nt (single-ended UMI) or 5-8nt (double-ended UMI). Its function is to freeze the state of DNA fragments before amplification, and each DNA molecule corresponds to a UMI. Therefore, during the analysis of bioinformatics, it can distinguish DNA templates from different sources, distinguish which are false-positive mutations caused by random errors in the process of PCR amplification and sequencing, and which are actually carried by patients, so as to filter out the background noise, realize the accurate detection of low-frequency and extremely low-frequency mutations, and carry out absolute quantification of different DNA molecules. It is widely used in low-frequency mutation detection, especially in the field of tumor research.

Fig 4 Schematic diagram of UMI adapter structure of Illumina platform
4.2 Complete adapters
Complete adapters, a necessary product for a PCR-free library, contain all the sequences required for sequencings, such as P5, P7, RdS1, and RdS2 in the Illumina platform, also index sequences and UMI sequences according to the requirements for sequencing. With the complete adapters, it can be directly sequenced without introducing other adapters through PCR. So, complete adapters can be used to build a PCR-free library. The PCR-free libraries can reduce PCR amplification bias, error rate, and sequence duplication, increasing the coverage of some high GC or high AT regions which are widely used in population genome research.
A complete adapter product for Illumina platform supplied by Yeasen (Cat#13519ES/13520ES)>>
A complete adapter product for MGI platform supplied by Yeasen(Cat#13360ES/13361ES)>>
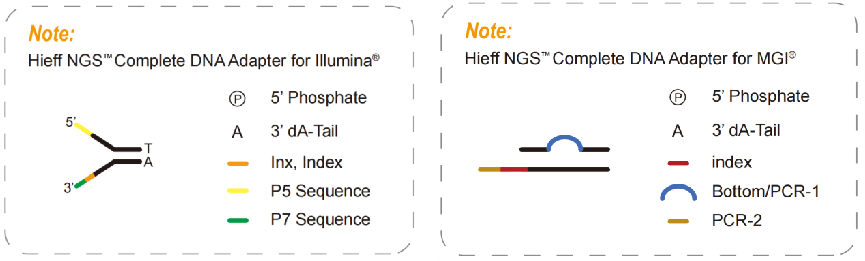
Fig. 5 Complete adapter diagram
4.3 Incomplete adapters
Incomplete adapters need to introduce other sequences by PCR after the adapter ligation to form a complete adapter. They are characterized by high connection efficiency and high effective library rate. The PCR process is an enrichment effect for the complete library to ensure the concentration of the effective library, and can also introduce double-ended indexes and UMI sequences.
4.4 Tn5 adapters
Tn5 adapters connect part of the adapter sequence to both ends of the DNA fragments through the restriction endonuclease activity of Tn5. They make fragmentation and adapter ligation carried out simultaneously to save time and samples. Finally, the rest of the linker sequence, index, UMI, and other sequences are introduced by PCR to form a complete library. It can be used to build a Cut&tag library.
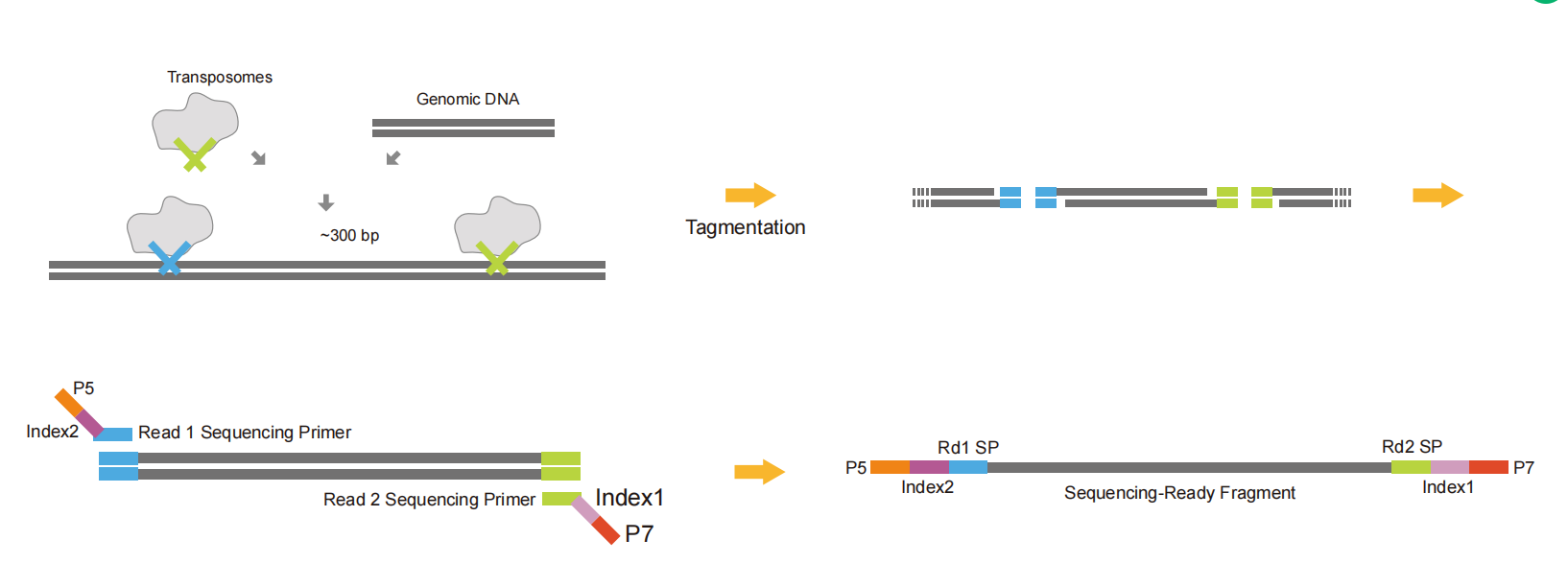
Fig.6 Schematic diagram of Tn5 adapter library construction
5. How to choose the right NGS adapters for your sequencing platforms?
Currently, there are two mainstream sequencing platforms, including Illumina and MGI. Yeasen, a complete solution for NGS provider, has developed multiple NGS adapters suitable to Illumina or MGI platforms.
In terms of the Illumina platform, Illumina NGS adapters supplied by Yeasen contain three types, including UDI, CDI, and a single index. In terms of the MGI platform, MGI NGS adapters offered by Yeasen have two types, involving Dual UMI -UDB and Single Index. We have listed the product information in the following table, including types of adapters, available sizes, and concentration of adapter and primer, respectively.
Complete and UDI NGS adapters do not need to worry about coupling problems, suitable for customers who want easy to use; CDI NGS adapters have fewer tubes and small size, which is suitable for customers who want to store and carry easily. PCR-free requires the use of complete NGS adapters.
| Platform | Name | Type | Cat# | Size | Describe | Concentration |
| Illumina | Hieff NGS™ Stubby UDI Primer Kit for Illumina | UDI | 12404ES/12405ES | 12×2 T/96×2 T/192×2 T/384×2 T | 12/96/192/384 kinds of index | Adapter:15μM; Primer:12.5μM |
| Hieff NGS™ 384 CDI Primer for Illumina, Set1-Set2 (Inquire) | CDI | 12412ES/12413ES | 96×2 T/96x20 T | 96 kinds of index | Adapter:15μM;Primer:25μM | |
| Hieff NGS™ RNA 384 CDI Primer for Illumina,Set1/Set2(Inquire) | RNA CDI | 12414ES/12415ES | 96×2 T/96x20 T | 96 kinds of index | Adapter:15μM;Primer:25μM | |
| Hieff NGS™ Complete Adapter Kit for Illumina, Set1/Set2 | Single Index (8bp) | 13519ES/13520ES | 48×4 T/48×16T | 96 kinds of index | 15μM | |
| MGI | Hieff NGS™ Dual UMI UDB Adapter Kit for MGI, Set1/Set2 | Dual UMI -UDB | 13367ES/13368ES | 48 x 2 T/48 x 4 T | 96 kinds of index | Adapter:10μM; Primer:12.5μM |
| Hieff NGS™ Complete Adapter Kit for MGI, Set1/Set2/Set3(Inquire) | Single Index | 13360ES | 8×2 T/8×4 T/8×100 T | 8 kinds of index, 41-48 | 10μM | |
| 13361ES | 16×2 T/16×4 T/16×100 T | 16 kinds of index, 57-72 | ||||
| 13362ES | 96×2 T/96×4 T/96×100 T | 96 kinds of index, 1-96 |
Regarding Reading
Key enzymes involved in NGS library construction
About NGS-related technology, how much do you know?
Various types of magnetic beads in NGS: DNA\RNA\mRNA magnetic beads
NGS library quantification: Quick and Accurate Qubit or Precise qPCR?All needed!