Transfection reagents have become routine reagents for studying gene function in eukaryotic cells. Transfection reagents are widely used in gene function research, gene expression regulation, mutation analysis, as well as gene therapy, cell therapy, protein production, and vaccine production.
For DNA transfection reagents and RNA transfection reagents, Yeasen has a strong R&D and production team, continuously optimizes formulas, improves production processes, and has launched a variety of products based on cationic liposomes and cationic polymers. Scientific research institutions and enterprises provide a full range of products, and the product line covers all fields involved in transfection reagents.
Comparison of different transfection methods
|
Transfection method |
Advantages |
Disadvantages |
|
Calcium Phosphate Coprecipitation |
Inexpensive and easy to operate |
The transfection efficiency is unstable, prone to DNA aggregation, etc., affecting the transfection efficiency, high cytotoxicity, and poor transfection efficiency |
|
Electroporation |
High efficiency for all cell types |
Electroporator is relatively expensive, with high cell lethality and large amount of nucleic acid and cells |
|
DEAE-dextran |
It is more effective for transfection of adherent cells, and can also be used for some suspension cells, with simple operation and reproducible results |
It has a preference for cells, has certain toxic and side effects on cells, and requires serum to inhibit cell growth during transfection |
|
ploybrene polybrene |
Relatively simple operation, average price |
Low efficiency and limited applicability |
|
Cationic liposome reagents |
Simple operation, high efficiency, wide applicability, good repeatability and low toxicity |
The price is slightly higher than other chemical reagents |
|
virus infection |
High efficiency, good transfection effect on difficult-to-transfect cells and primary cells |
The virus packaging process is complicated and the virus is dangerous |
|
Polyethylenimine (PEI) |
Low price, simple operation, wide application range |
Less efficient than liposome-based transfection reagents, some cells are sensitive to contact |
As a cationic liposome transfection reagent, Hieff Trans™ liposome nucleic acid transfection reagent has higher transformation efficiency and more convenient operation than other transfection reagent. It has been validated in a variety of cell lines, see the summary of validated cell lines below.
The principle of action
Cationic liposomes can encapsulate nucleic acids through the electrostatic interaction of positive charges on the surface to form nucleic acid-liposome complexes. The surface of the cell membrane is negatively charged, which can adsorb the complex, and the adsorbed complex can form an inclusion body into the cell through membrane fusion or endocytosis. A small part of DNA can be released from inclusion bodies and enter the cytoplasm, and then further enter the nucleus for transcription and expression.
Features of Hieff Trans™ Transfection Reagent
Efficiency: Most eukaryotic cells can be transfected, and the transfection efficiency for common cell lines is over 90%;
Low toxicity: the transfected cells are in good shape and express a large amount of transfected gene proteins;
Easy to operate: the liposome complex can be directly added to the serum-containing medium;
Wide applicability: suitable for transient transfection and stable transfection experiments.
Operating procedures
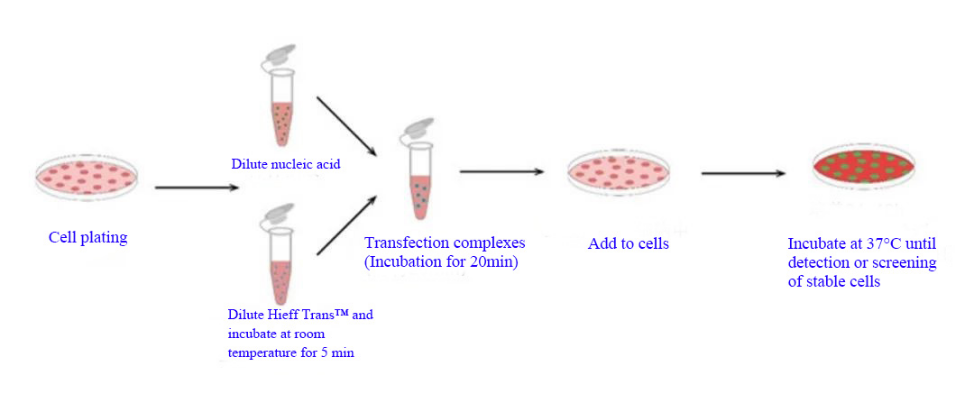
Figure 1. Hieff Trans™ Transfection Reagent operation flow map
Hieff Trans™ is supplied in sterile liquid form. Generally, for 24-well plate transfection, about 1.5 μL each time, 1 mL Hieff Trans™ can do about 660 transfections; for 6-well plate, about 6 μL each time, 1 mL Hieff Trans™ can do about 160 transfections.
Product data
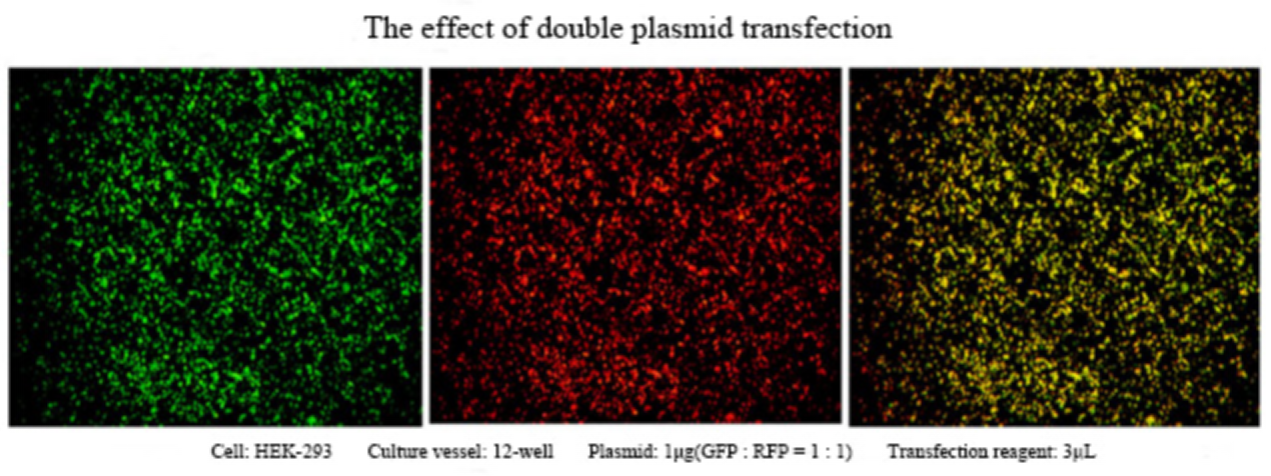
Figure 2. The effect of double plasmid transfection
Validated cell line summary
| 293T | 293FT | HEK293 | HEK 293T | Caco2 |
| COS-7 | DF-1 | Lenti X-293T | HeLa | HepG2 |
| LM3 | MCF-7 | MDA-MB-231 | MEF | NCI-H1975 |
| NIH-3T3 | PC12 | Raw264.7 | Vero | H9c2 |
| Hepa1-6 | MCF10A | CHO | 5-8F | BV-2 |
| A549 | 3t3 | H520 | HUVEC | C6 |
| HaCaT | N2A | SGC-7901 | H9 | MKN-28 |
| Hep 3B | SMCC7721 | RKO | C2C12 | More… |
Partial cell line transfection data provided by customers (for reference only)
|
Cell |
Culture vessel |
Cell density |
DNA |
Hieff Trans™ |
Efficiency |
|
293T |
6-well |
80% |
1μg |
2μL |
90% |
|
293FT |
2-well |
85% |
1μg |
4μL |
90% |
|
Hela |
12-well |
90% |
0.2μg |
0.6μL |
90% |
|
Raw264.7 |
35mm |
80% |
1μg |
2μL |
90% |
|
Hek293 |
6-well |
95% |
2μg |
10μL |
80-90% |
|
NCI-H1975 |
6-well |
80% |
4μg |
10μL |
80% |
|
3t3 |
12-well |
90% |
1μg |
5μL |
50% |
Price comparison
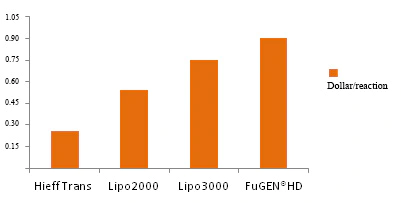
Figure 3. Price comparison of each company for a single transfection reaction
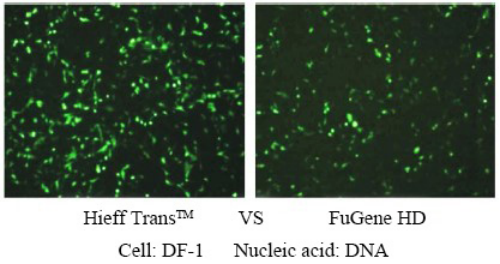
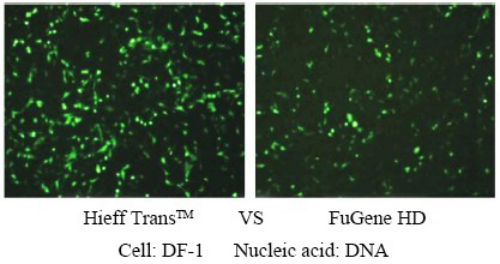
Figure 4. Comparison of the effects of different reagents
Products information
| Product name | SKU | Specifications |
| Hieff Trans™ Liposomal Transfection Reagent | 40802ES02 | 0.5 mL |
| 40802ES03 | 1.0 mL | |
| 40802ES08 | 5×1mL | |
| Hieff Trans™ Suspension Cell-Free Liposomal Transfection Reagent (Inquire) | 40805ES02 | 0.5 mL |
| 40805ES03 | 1.0 mL | |
| 40805ES08 | 5×1 mL | |
| Hieff Trans™ in vitro siRNA/miRNA Transfection Reagent (Inquire) | 40806ES02 | 0.5 mL |
| 40806ES03 | 1.0 mL | |
| Polyethylenimine Linear(PEI) MW40000(rapid lysis) | 40816ES02 | 100 mg |
| 40816ES03 | 1 g | |
| 40816ES08 | 5×1 g |
Some of the articles published using our products
[1] Liu R, Yang J, Yao J, Zhao Z, He W, Su N, Zhang Z, Zhang C, Zhang Z, Cai H, Zhu L, Zhao Y, Quan S, Chen X, Yang Y. Optogenetic control of RNA function and metabolism using engineered light-switchable RNA-binding proteins. Nat Biotechnol. 2022 Jan 3. doi: 10.1038/s41587-021-01112-1. Epub ahead of print. PMID: 34980910. (IF:54.908)
[2] Zhou J, Chen P, Wang H, Liu H, Li Y, Zhang Y, Wu Y, Paek C, Sun Z, Lei J, Yin L. Cas12a variants designed for lower genome-wide off-target effect through stringent PAM recognition. Mol Ther. 2022 Jan 5;30(1):244-255. doi: 10.1016/j.ymthe.2021.10.010. Epub 2021 Oct 20. PMID: 34687846; PMCID: PMC8753454. (IF:11.454)
[3] Chen S, Cao X, Zhang J, Wu W, Zhang B, Zhao F. circVAMP3 Drives CAPRIN1 Phase Separation and Inhibits Hepatocellular Carcinoma by Suppressing c-Myc Translation. Adv Sci (Weinh). 2022 Mar;9(8):e2103817. doi: 10.1002/advs.202103817. Epub 2022 Jan 24. PMID: 35072355; PMCID: PMC8922094. (IF:16.808)
[4] Zhang Y, Yu X, Sun R, Min J, Tang X, Lin Z, Xie S, Li X, Lu S, Tian Z, Gu C, Teng L, Yang Y. Splicing factor arginine/serine-rich 8 promotes multiple myeloma malignancy and bone lesion through alternative splicing of CACYBP and exosome-based cellular communication. Clin Transl Med. 2022 Feb;12(2):e684. doi: 10.1002/ctm2.684. PMID: 35184390. (IF:11.492)
[5] Tang X, Deng Z, Ding P, Qiang W, Lu Y, Gao S, Hu Y, Yang Y, Du J, Gu C. A novel protein encoded by circHNRNPU promotes multiple myeloma progression by regulating the bone marrow microenvironment and alternative splicing. J Exp Clin Cancer Res. 2022 Mar 8;41(1):85. doi: 10.1186/s13046-022-02276-7. PMID: 35260179. (IF:11.161)
[6] Hua Z, Wei R, Guo M, Lin Z, Yu X, Li X, Gu C, Yang Y. YTHDF2 promotes multiple myeloma cell proliferation via STAT5A/MAP2K2/p-ERK axis. Oncogene. 2022 Mar;41(10):1482-1491. doi: 10.1038/s41388-022-02191-3. Epub 2022 Jan 24. PMID: 35075244. (IF:9.867)
[7] Liang Y, Lu Q, Li W, Zhang D, Zhang F, Zou Q, Chen L, Tong Y, Liu M, Wang S, Li W, Ren X, Xu P, Yang Z, Dong S, Zhang B, Huang Y, Li D, Wang H, Yu W. Reactivation of tumour suppressor in breast cancer by enhancer switching through NamiRNA network. Nucleic Acids Res. 2021 Sep 7;49(15):8556-8572. doi: 10.1093/nar/gkab626. PMID: 34329471; PMCID: PMC8421228. (IF:16.9)
[8] Dai L, Dai Y, Han J, Huang Y, Wang L, Huang J, Zhou Z. Structural insight into BRCA1-BARD1 complex recruitment to damaged chromatin. Mol Cell. 2021 Jul 1;81(13):2765-2777.e6. doi: 10.1016/j.molcel.2021.05.010. Epub 2021 Jun 7. PMID: 34102105. (IF:17.97)
[9] Zhang K, Wang A, Zhong K, Qi S, Wei C, Shu X, Tu WY, Xu W, Xia C, Xiao Y, Chen A, Bai L, Zhang J, Luo B, Wang W, Shen C. UBQLN2-HSP70 axis reduces poly-Gly-Ala aggregates and alleviates behavioral defects in the C9ORF72 animal model. Neuron. 2021 Jun 16;109(12):1949-1962.e6. doi: 10.1016/j.neuron.2021.04.023. Epub 2021 May 14. PMID: 33991504. (IF:17.17)
[10] Liang Y, Lu Q, Li W, Zhang D, Zhang F, Zou Q, Chen L, Tong Y, Liu M, Wang S, Li W, Ren X, Xu P, Yang Z, Dong S, Zhang B, Huang Y, Li D, Wang H, Yu W. Reactivation of tumour suppressor in breast cancer by enhancer switching through NamiRNA network. Nucleic Acids Res. 2021 Sep 7;49(15):8556-8572. doi: 10.1093/nar/gkab626. PMID: 34329471; PMCID: PMC8421228. (IF:16.9)
[11] Li T, Chen X, Qian Y, Shao J, Li X, Liu S, Zhu L, Zhao Y, Ye H, Yang Y. A synthetic BRET-based optogenetic device for pulsatile transgene expression enabling glucose homeostasis in mice. Nat Commun. 2021 Jan 27;12(1):615. doi: 10.1038/s41467-021-20913-1. PMID: 33504786; PMCID: PMC7840992. (IF:14.92)
[12] Pan Y, He X, Li C, Li Y, Li W, Zhang H, Wang Y, Zhou G, Yang J, Li J, Qu J, Wang H, Gao Z, Shen Y, Li T, Hu H, Ma H. Neuronal activity recruits the CRTC1/CREB axis to drive transcription-dependent autophagy for maintaining late-phase LTD. Cell Rep. 2021 Jul 20;36(3):109398. doi: 10.1016/j.celrep.2021.109398. PMID: 34289350. (IF:9.42)
[13] Liu H, Xing R, Ou Z, Zhao J, Hong G, Zhao TJ, Han Y, Chen Y. G-protein-coupled receptor GPR17 inhibits glioma development by increasing polycomb repressive complex 1-mediated ROS production. Cell Death Dis. 2021 Jun 12;12(6):610. doi: 10.1038/s41419-021-03897-0. PMID: 34120140; PMCID: PMC8197764. (IF:8.463)
[14] Fan Y, Wang J, Jin W, Sun Y, Xu Y, Wang Y, Liang X, Su D. CircNR3C2 promotes HRD1-mediated tumor-suppressive effect via sponging miR-513a-3p in triple-negative breast cancer. Mol Cancer. 2021 Feb 2;20(1):25. doi: 10.1186/s12943-021-01321-x. PMID: 33530981; PMCID: PMC7851937. (IF:27.403)
[15] Gu C, Wang Y, Zhang L, Qiao L, Sun S, Shao M, Tang X, Ding P, Tang C, Cao Y, Zhou Y, Guo M, Wei R, Li N, Xiao Y, Duan J, Yang Y. AHSA1 is a promising therapeutic target for cellular proliferation and proteasome inhibitor resistance in multiple myeloma. J Exp Clin Cancer Res. 2022 Jan 6;41(1):11. doi: 10.1186/s13046-021-02220-1. PMID: 34991674; PMCID: PMC8734095. (IF:11.161)
[16] Luo Q, Wu X, Zhao P, Nan Y, Chang W, Zhu X, Su D, Liu Z. OTUD1 Activates Caspase-Independent and Caspase-Dependent Apoptosis by Promoting AIF Nuclear Translocation and MCL1 Degradation. Adv Sci (Weinh). 2021 Feb 8;8(8):2002874. doi: 10.1002/advs.202002874. PMID: 33898171; PMCID: PMC8061361. (IF:15.84)
[17] Luo Q, Wu X, Nan Y, Chang W, Zhao P, Zhang Y, Su D, Liu Z. TRIM32/USP11 Balances ARID1A Stability and the Oncogenic/Tumor-Suppressive Status of Squamous Cell Carcinoma. Cell Rep. 2020 Jan 7;30(1):98-111.e5. doi: 10.1016/j.celrep.2019.12.017. PMID: 31914402. (IF:9.42)
[18] Sun X, Peng X, Cao Y, Zhou Y, Sun Y. ADNP promotes neural differentiation by modulating Wnt/β-catenin signaling. Nat Commun. 2020 Jun 12;11(1):2984. doi: 10.1038/s41467-020-16799-0. PMID: 32533114; PMCID: PMC7293280. (IF:14.911)
[19] Yang X, Wang H, Xie E, Tang B, Mu Q, Song Z, Chen J, Wang F, Min J. Rewiring ERBB3 and ERK signaling confers resistance to FGFR1 inhibition in gastrointestinal cancer harbored an ERBB3-E928G mutation. Protein Cell. 2020 Dec;11(12):915-920. doi: 10.1007/s13238-020-00749-z. PMID: 32632529; PMCID: PMC7719122. (IF:14.872)
[20] Chen, T., Chen, Y., Chen, H. et al. Dual-enzyme-propelled unbounded DNA walking nanomachine for intracellular imaging of lowly expressed microRNA. Nano Res. 12, 1055–1060 (2019). https://doi.org/10.1007/s12274-019-2344-5 (IF:8.21)
[21] Zhang X, Qi Z, Yin H, Yang G. Interaction between p53 and Ras signaling controls cisplatin resistance via HDAC4- and HIF-1α-mediated regulation of apoptosis and autophagy. Theranostics. 2019 Jan 30;9(4):1096-1114. doi: 10.7150/thno.29673. PMID: 30867818; PMCID: PMC6401400. (IF:8.12)
[22] Zou Y, Wang A, Shi M, Chen X, Liu R, Li T, Zhang C, Zhang Z, Zhu L, Ju Z, Loscalzo J, Yang Y, Zhao Y. Analysis of redox landscapes and dynamics in living cells and in vivo using genetically encoded fluorescent sensors. Nat Protoc. 2018 Oct;13(10):2362-2386. doi: 10.1038/s41596-018-0042-5. PMID: 30258175; PMCID: PMC6714056. (IF:13.49)
[23] Zhang K, Zhao X, Chen X, Wei Y, Du W, Wang Y, Liu L, Zhao W, Han Z, Kong D, Zhao Q, Guo Z, Han Z, Liu N, Ma F, Li Z. Enhanced Therapeutic Effects of Mesenchymal Stem Cell-Derived Exosomes with an Injectable Hydrogel for Hindlimb Ischemia Treatment. ACS Appl Mater Interfaces. 2018 Sep 12;10(36):30081-30091. doi: 10.1021/acsami.8b08449. Epub 2018 Aug 29. PMID: 30118197. (IF:8.09)
[24] Hao H, Hu S, Chen H, Bu D, Zhu L, Xu C, Chu F, Huo X, Tang Y, Sun X, Ding BS, Liu DP, Hu S, Wang M. Loss of Endothelial CXCR7 Impairs Vascular Homeostasis and Cardiac Remodeling After Myocardial Infarction: Implications for Cardiovascular Drug Discovery. Circulation. 2017 Mar 28;135(13):1253-1264. doi: 10.1161/CIRCULATIONAHA.116.023027. Epub 2017 Feb 2. PMID: 28154007. (IF:18.881)