Hieff NGS™ DNA Selection Beads
Description
Hieff NGS™ DNA Selection Beads are prepared based on the SPRI (Solid Phase Reverse Immobilization) principle and is applicable for DNA purification and size selection during the preparation of next generation sequencing (NGS) libraries. Hieff NGS™ DNA Selection Beads is compatible with various of DNA and RNA library prep kits.
Components
|
Components No. |
Name |
12601ES08 |
12601ES56 |
12601ES75 |
|
12601 |
Hieff NGS™ DNA Selection Beads |
5 mL |
60 mL |
450 mL |
Specifications
|
Product Line |
DNA clean and selection Beads |
|
Starting Material |
DNA |
|
Compatibility |
DNA |
|
Isolation Technology |
Magnetic Bead |
|
Final Product Type |
DNA |
|
For Use With (Application) |
DNA celan up, DNA size slection |
Shipping and Storage
The beads are shipping with ice packs and can be stored at 2-8°C for 18 month.
Instructions
- Preparation
Equilibrate the selection beads at room temperature for at least 30 min before use.
- Size selection
The operation flow of size selection is shown in Figure 1 and the protocol is as follows.
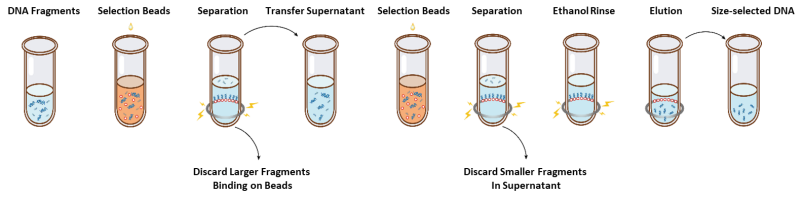
Figure 1. The Operation Flowchart of Size Selection
2.1 Mix the beads thoroughly by vortexing or pipetting up and down every time before using.2.2 Add the first round of selection beads to the sample (refer to Table 1). Mix thoroughly by vortexing or pipetting up and down at least 10 times.
2.3 Incubate at room temperature for 5 min.
2.4 Spin down the tube briefly and place it on magnetic stand. When the solution is clear (about 5 min), transfer the supernatant to a new PCR tube.
2.5 Add the second round of selection beads to the sample from step 2.4 according to Table 1. Mix thoroughly by vortexing or pipetting up and down at least 10 times.
2.6 Incubate at room temperature for 5 min.
2.7 Spin down the tube briefly and place it on magnetic stand. When the solution is clear (about 5 min), aspirate the supernatant and discard.
2.8 Keep the tube in the magnetic stand and add 200 μL of freshly prepared 80% ethanol to without disturbing the beads, incubate at room temperature for 30 sec. Aspirate the ethanol and discard.
2.9 Repeat step 2.8 once for a total of two washes.
2.10 Remove residual ethanol with 10 µL pipette tips. Keep the tube in the magnetic stand, air dry the selection beads with the lid open until cracks just appear (about 5 min).
Note: Do not over-dry the selection beads. This may result in lower recovery DNA target.
2.11 Remove the tube from the magnetic stand. Add an appropriate amount of ddH2O (≥20 µL) and mix thoroughly by vortexing or pipetting up and down at least 10 times.
2.12 Incubate at room temperature for 5 min.
2.13 Spin down the tube briefly and place it on the magnetic stand. When the solution is clear (about 5 minutes), transfer 20 μL of the supernatant to a new tube.
- Recommended Conditions for DNA Size Selection
The calf thymus DNA was fragmented by sonication to prepare a fragment of 100-1,000 bp, and two rounds of size selection were performed according to Table 1. The results were analyzed using Agilent 2100 Bioanalyzer (Figure 2).
Table 1. Recommended condition for DNA size selection
|
Length of DNA fragment |
250-350 bp |
320-420 bp |
450-550 bp |
550-700 bp |
700-900 bp |
800-1,000 bp |
|
Ratio of Beads: DNA for the 1st Round |
0.80× |
0.70× |
0.60× |
0.55× |
0.50× |
0.45× |
|
Ratio of Beads: DNA for the 2nd Round |
0.20× |
0.20× |
0.20× |
0.15× |
0.15× |
0.15× |
Note: "×" in the table indicates the volume of sample DNA. For example, if the insert length of the library is 250 bp and the sample DNA volume is 100 μL, the volume of magnetic beads used in the first round of sorting is 0.80×100 μL=80 μL; the volume of magnetic beads used in the second round of sorting is 0.20× 100 μL=20 μL.
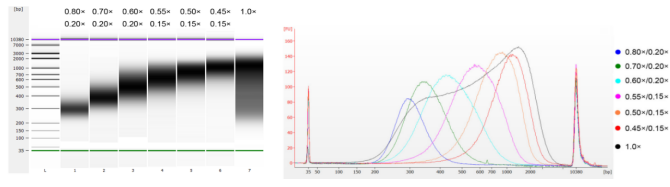
Figure 2. Agilent 2100 high sensitivity DNA chip electropherogram
Notes:
1. For your safety and health, please wear lab coats and disposable gloves for operation.
[1] Wang X, Yuan Q, Zhang W, et al. Sequence specific integration by the family 1 casposase from Candidatus Nitrosopumilus koreensis AR1. Nucleic Acids Res. 2021;49(17):9938-9952. doi:10.1093/nar/gkab725(IF:16.971)
[2] Duan XZ, Sun JT, Wang LT, et al. Recent infection by Wolbachia alters microbial communities in wild Laodelphax striatellus populations. Microbiome. 2020;8(1):104. Published 2020 Jul 2. doi:10.1186/s40168-020-00878-x(IF:11.607)
[3] Song B, Almatrafi E, Sang F, et al. Managing Fenton-treated sediment with biochar and sheep manure compost: Effects on the evolutionary characteristics of bacterial community. J Environ Manage. 2022;316:115218. doi:10.1016/j.jenvman.2022.115218(IF:6.789)
[4] Huang C, Mei Q, Lou L, et al. Ulcerative Colitis in Response to Fecal Microbiota Transplantation via Modulation of Gut Microbiota and Th17/Treg Cell Balance. Cells. 2022;11(11):1851. Published 2022 Jun 5. doi:10.3390/cells11111851(IF:6.600)
[5] Ghosh S, Yang X, Wang L, Zhang C, Zhao L. Active phase prebiotic feeding alters gut microbiota, induces weight-independent alleviation of hepatic steatosis and serum cholesterol in high-fat diet-fed mice. Comput Struct Biotechnol J. 2020;19:448-458. Published 2020 Dec 24. doi:10.1016/j.csbj.2020.12.011(IF:6.018)
[6] Gao X, Yu B, Yu J, et al. Developmental Profiling of Dietary Carbohydrate Digestion in Piglets. Front Microbiol. 2022;13:896660. Published 2022 Apr 29. doi:10.3389/fmicb.2022.896660(IF:5.640)
[7] Li P, Zhang Y, Yan F, Zhou X. Characteristics of a Bacteriophage, vB_Kox_ZX8, Isolated From Clinical Klebsiella oxytoca and Its Therapeutic Effect on Mice Bacteremia. Front Microbiol. 2021;12:763136. Published 2021 Dec 3. doi:10.3389/fmicb.2021.763136(IF:5.640)
[8] Lin Z, Luo P, Huang D, Wu Y, Li F, Liu H. Multi-omics based strategy for toxicity analysis of acrylamide in Saccharomyces cerevisiae model. Chem Biol Interact. 2021;349:109682. doi:10.1016/j.cbi.2021.109682(IF:5.194)
[9] Sun X, Lv W, Wang Y, et al. Mrgprb2 gene plays a role in the anaphylactoid reactions induced by Houttuynia cordata injection. J Ethnopharmacol. 2022;289:115053. doi:10.1016/j.jep.2022.115053(IF:4.360)
[10] Ma H, Lai B, Zan C, Di X, Zhu X, Wang K. GLO1 Contributes to the Drug Resistance of Escherichia coli Through Inducing PER Type of Extended-Spectrum β-Lactamases. Infect Drug Resist. 2022;15:1573-1586. Published 2022 Apr 5. doi:10.2147/IDR.S358578(IF:4.003)
[11] Zhong Y, Zhao W, Tang Z, et al. Comparative transcriptomic analysis of the different developmental stages of ovary in red swamp crayfish Procambarus clarkii. BMC Genomics. 2021;22(1):199. Published 2021 Mar 21. doi:10.1186/s12864-021-07537-x(IF:3.969)
[12] Lian C, Yang H, Lan J, et al. Comparative analysis of chloroplast genomes reveals phylogenetic relationships and intraspecific variation in the medicinal plant Isodon rubescens. PLoS One. 2022;17(4):e0266546. Published 2022 Apr 6. doi:10.1371/journal.pone.0266546(IF:3.240)
[13] Diao G, Huang J, Zheng X, et al. Prostaglandin E2 serves a dual role in regulating the migration of dendritic cells. Int J Mol Med. 2021;47(1):207-218. doi:10.3892/ijmm.2020.4801(IF:3.098)
[14] Bing XL, Zhao DS, Peng CW, Huang HJ, Hong XY. Similarities and spatial variations of bacterial and fungal communities in field rice planthopper (Hemiptera: Delphacidae) populations. Insect Sci. 2020;27(5):947-963. doi:10.1111/1744-7917.12782(IF:2.791)
[15] Li X, Zhou S, Zhang J, Zhou Z, Xiong Q. Directional Changes in the Intestinal Bacterial Community in Black Soldier Fly (Hermetia illucens) Larvae. Animals (Basel). 2021;11(12):3475. Published 2021 Dec 6. doi:10.3390/ani11123475(IF:2.752)
[16] Yang J, Peng Y, Kong W. Identification and complete genome sequence of mulberry cryptic virus 1. Arch Virol. 2022;167(2):687-690. doi:10.1007/s00705-021-05350-1(IF:2.574)
[17] Chang Y, Xia X, Sui L, et al. Endophytic colonization of entomopathogenic fungi increases plant disease resistance by changing the endophytic bacterial community. J Basic Microbiol. 2021;61(12):1098-1112. doi:10.1002/jobm.202100494(IF:2.281)
[18] Ding CY, Ma YM, Li B, et al. Identification and Functional Analysis of Differentially Expressed Genes in Myzus persicae (Hemiptera: Aphididae) in Response to Trans-anethole. J Insect Sci. 2022;22(1):3. doi:10.1093/jisesa/ieab094(IF:1.857)
Catalog No.:*
Name*
phone Number:*
Lot:*
Email*
Country:*
Company/Institute:*

